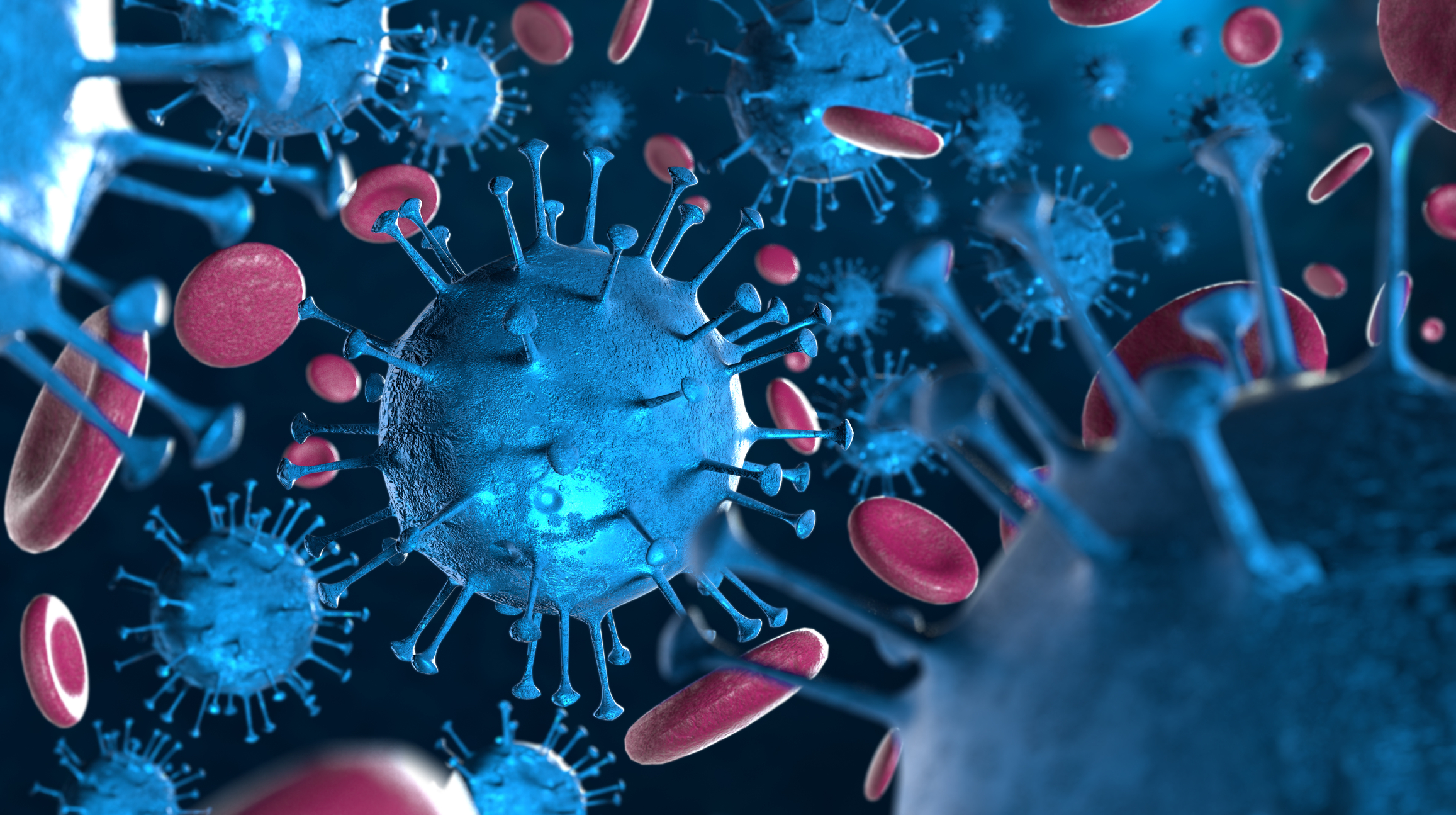
COVID-19, a highly contagious respiratory illness, originates from enveloped coronavirus virions characterized by distinctive, spherical-shaped crown spikes on their surface.
COVID-19: A Race Against Time – Understanding Pandemic Speed
The disease COVID-19 is caused by Coronavirus virions that are enveloped spherical-shaped virus particles with a diameter ranging from 80 to 160 nanometres (625 smaller than a single strand width of a hair). These virus particles contain surface ‘crown’ projections of up to 20 nm in length, covering the entire virion surface. Since viruses do not have metabolic systems and are believed to be non-living, according to some researchers, they act as intracellular parasites. They transfer RNA or DNA genomes into the host from their protective, virus-coded symmetric protein capsid: the nucleoprotein and the genome form the nucleocapsid. In enveloped viruses, the nucleocapsid is coated by a lipid bilayer which results from the modified host cell membrane and is covered with an outer layer of the viral envelope of glycoproteins.
The coronaviruses belong to the order Nidovirales of viruses with animal and human hosts and include the families Coronaviridae, Arteriviridae, Roniviridae and Mesoniviridae. Coronaviruses are classified into four genera:
The alphacoronavirus include porcine respiratory coronavirus (PRCV), porcine epidemic diarrhoea virus (PEDV), human coronavirus NL63 (HCoV-NL63) and porcine transmissible gastroenteritis coronavirus (TGEV).
The betacoronaviruses include human coronavirus OC43, Middle East respiratory syndrome-related coronavirus (MERS-CoV), Severe Acute Respiratory Syndrome-associated coronavirus (SARS-CoV), mouse hepatitis coronavirus (MHV), bovine coronavirus (BCoV) and bat coronavirus HKU4.
All these betacoronaviruses infect mammals. The gamma coronaviruses infect avian species and include avian infectious bronchitis coronavirus (IBV) and deltacoronaviruses such as porcine deltacoronavirus which infect both mammalian and avian species.
Inside the coronavirus, the envelope is a helical nucleocapsid of 6-8 nm in diameter. The formation of a helical nucleocapsid in coronavirus is unexpected because a helical nucleocapsid is usually associated with viruses containing a negative-stranded RNA genome. However, coronavirus contains a positive-stranded RNA (positive-sense) genome. In most cases, the positive-stranded RNA viruses have icosahedral nucleocapsids and may play a role in the mechanism of coronavirus RNA synthesis.
The positive-sense viral RNA genome (Group IV in the Baltimore) can facilitate messenger RNA and thus can be translated into protein in the host cell. Positive-strand RNA viruses encompass over 33% of all virus genera. They include numerous pathogens such as West Nile, dengue, and hepacivirus C (HCV) and the coronaviruses SARS, MERS and SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2). In addition, a positive sense of viral RNA is present in the rhinoviruses that cause the common cold.
Note: The official names for the virus responsible for COVID-19 (previously known as 2019 novel coronavirus, 2019-nCoV) and the disease it causes is:
Coronavirus disease (COVID-19); Virus: severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
All coronavirus particles contain three to four structural proteins; the first is a spike protein (S) shaped like a club. This glycoprotein spike (peplomer) is formed on a viral capsid or viral envelope and has a molecular weight of 180 kilodaltons. The function of the spike protein is to mediate the coronavirus entry into host cells. The binding to a receptor on the host cell surface is facilitated through the S1 subunit and initiates the fusion of the viral and host membranes via its S2 subunit. The coronavirus family have two domains in S1 that are different and can recognise a range of host receptors leading to the attachment of the viral particle. Subsequently, the spike protein can exist in two conformations which are the prefusion and post-fusion. The conformational change from prefusion to post fusion of the spike protein will trigger membrane fusion.
Coronaviruses have club-shaped spikes on their outer coats. Immune responses from other coronavirus studies suggest that these are a good target for a vaccine.
The second viral structural protein, M, is also an integral membrane glycoprotein. The function of this protein is unclear: it is unlike the matrix protein present in other enveloped viruses. The inside domain of the M protein may interact with the virion nucleocapsid. This interaction could be the focal point for the assembly of virus particles because virus budding appears to occur at the site of M accumulation. The monoclonal antibodies specific to the M protein do not neutralise virus infectivity. The third glycoprotein on the virion surface is HE. This protein most likely constitutes the smaller spikes observed on virus particles in some electron micrographs of coronaviruses. The fourth structural protein is an internal component of the virus. This protein, N, is a phosphoprotein of 50 kilodaltons, constituting the virus’s nucleocapsid protein. The protein binds to virion RNA, providing the structural basis for the helical nucleocapsid.
Coronavirus contains a single piece of non-segmented RNA genome with an estimated molecular weight of 6-8 megadaltons. The RNA contains a 5′ cap structure and 3′ poly(A) tail and is infectious upon transfection (the process of delivering nucleic acids and small proteins into eukaryotic cells) of the naked RNA into a susceptible cell line. The 5′ cap protects the mRNA from degradation and facilitates ribosome binding during translation. A poly (A) tail is added to the 3′ end of the pre-mRNA once elongation is complete.
The RNA also serves as a template for in vitro translation of viral proteins. Thus, this RNA is typically positive-stranded (sense-strand RNA virus). In these cases, the virus’s genetic information comprises a single strand of RNA that is the positive (sense) strand that encodes messenger RNA and protein. Replication in positive-strand RNA viruses is via a negative-strand intermediate. Examples of positive-strand RNA viruses include poliovirus, Coxsackie virus and echovirus.
No negative-stranded RNA has been detected in the virion for coronavirus. The RNA is more significant than any other known viral RNA (the next smaller viral RNA is paramyxovirus RNA, which is approximately 14 kilodaltons long).
Coronaviruses generally have very restricted host ranges, infecting only cells of their host species. However, some cross-species infections do occur. For instance, BCV can infect bovine, human, and rat cells. In 1965, Tyrrell and Bynoe discovered the first human coronavirus (HCoV) called B814. They isolated the virus from the respiratory tract of an adult with a common cold. However, Tyrrell and Bynoe could not grow the agent in tissue culture, and eventually, Hamre and Procknow successfully grew the virus.
From Ground Zero to Worldwide Turmoil: The Emergence of the Coronavirus Outbreak
In December 2019, Wuhan, the capital of Central China’s Hubei province – a population of over 18 million people, became the epicentre for an outbreak of pneumonia of unknown origin. At the time, this outbreak was reported to the WHO (World Health Organisation) and the Chinese health authorities, who implemented an investigation to identify the source of infection and to control the disease. These measures involved isolating people suspected to have the disease and closely monitoring contacts. Further measures included collecting and analysing epidemiological and clinical data from patients, including developing diagnostic and treatment plans to address this outbreak.
Then on 7 January 2020, Chinese scientists isolated a novel coronavirus (CoV) from patients in Wuhan. The patients were experiencing symptoms similar to those caused by severe acute respiratory syndrome coronavirus (SARS-CoV). Since the SARS outbreak in 2002, investigations have continued into the interaction of SARS-CoV spike protein receptor-binding domain (RBD) with the host receptor angiotensin-converting enzyme 2 (ACE2).
This interaction between the two receptors regulates the cross-species and human-to-human transmissions of SARS-CoV.
Therefore, the analyses of the potential receptor usage by SARS-CoV-2 is based on knowledge about SARS-CoV and the newly released sequence of 2019-nCoV.
Coronavirus Uncovered: Understanding the Distinctive Properties and Behavior
- The sequence of the 2019-nCoV receptor-binding domain identified the interaction between the receptor-binding motif (RBM) and ACE2. This interaction is similar to that of SARS-CoV, and evidence suggests that 2019-nCoV uses ACE2 as its receptor.
- Several amino acid residues in 2019-nCoV RBM, such as Gln493, provide favourable interactions with human ACE2. Therefore, this observation is consistent with the 2019-nCoV ability to infect human cells.
- Also, the amino acid residue Asn501 in 2019-nCoV RBM unusually binds human ACE2. This suggests that 2019-nCoV has acquired some ability for human-to-human transmission.
- However, phylogenetic analysis indicates a bat origin of 2019-nCoV, but 2019-nCoV potentially recognises ACE2 from a diverse range of animal species except for mice and rats. Therefore, this implicates these animal species as possible intermediate hosts or animal models for 2019-nCoV infections.
- These analyses provide insights into the receptor usage, cell entry, host cell infectivity and animal origin of 2019-nCoV and may help epidemic surveillance and preventive measures against 2019-nCoV.
The genetic sequence (GenBank: MN908947.3) of the 2019 novel coronavirus (2019-nCoV) enabled the rapid development of point-of-care real-time polymerase chain reaction (PCR) diagnostic tests specific for 2019-nCoV. These tests were based on full genome sequence data. The analysis of all cases diagnosed as of 11 February 2020, by using China’s Infectious Disease Information System gave the following results from a total of 72314 patient records: 44672 (61.8%) confirmed cases of COVID-19 and 16,186 (22.4%) suspected COVID-19 cases, in addition to, 10567 (14.6%) clinically diagnosed cases (Hubei only) and 889 asymptomatic cases (1.2%)-contributed data for the analysis.
The clinical features of the first 41 patients (see table below) admitted to a hospital in Wuhan were confirmed to be infected with 2019-nCoV by January 2020. The symptoms resulting from the 2019-nCoV infection were non-specific, including fever, dry cough and sickness.
Characteristics of patients infected with SARS-CoV-2, MERS-CoV, and SARS-CoV:
Symptoms of COVID-19
COVID-19 Radiographic Features: Unraveling Atypical Pneumonia through Chest Radiographs and CT Imaging
The radiographic features of COVID-19 using chest radiographs and Computed Tomography (CT) are those of atypical pneumonia. However, imaging has limited sensitivity for COVID-19, and up to 18% demonstrated normal chest radiographs or CT subjected to mild or early disease course. This decreases to 3% in severe diseases. Bilateral and multilobar involvement is frequent. The application of a plain radiograph is less sensitive than a chest CT. However, chest radiography is usually the first-line imaging modality for patients with suspected COVID-19. The chest radiographs may be normal in early or mild disease. Of patients with COVID-19 requiring hospitalisation, 69% of initial admissions gave an abnormal chest radiograph. Also, 80% of hospital patients indicated some form of radiographic abnormalities. These observations were found to be more prominent after 10-12 days from the onset of symptoms. The most common findings were airspace opacities compared to the less frequent pulmonary ground-glass opacity. The distribution of COVID-19 in the chest is bilateral, and peripheral, with the lower region being predominant. This is in contrast to multiple lung parenchymal abnormalities and associated pleural effusion, which is rare.
Decoding COVID-19 in Adults: The Role of Computed Tomography (CT) Imaging
As COVID-19 spreads, efforts are being made to reduce transmission via social distancing, isolation of cases and tracing of contacts. Therefore, the current COVID-19 situation requires the development of a vaccine or antiviral drugs. Clinical trials of hydroxychloroquine already used in treating malaria, lupus erythematosus and rheumatoid arthritis are under investigation for COVID-19 pneumonia in China (NCT04261517 and NCT04307693). The first study (NCT04261517) has shown positive preliminary outcomes in terms of clinical management.
Scientists continue to investigate how the COVID-19 virus passed to humans in the first place. Even though bats are thought to be the virus’s origin, bat coronaviruses differ from COVID-19. For example, the spike proteins, which bind to receptors on the surface of cells to gain access, are different in the two viruses. It was found that coronavirus RNA – isolated from pangolins encoded – spike proteins that were similar to COVID-19. However, that does not prove that the new coronavirus passed through pangolins.
Unveiling the Secrets: Exploring the Mechanism of SARS-CoV-2 Infection
The betacoronavirus genome can encode the glycosylated spike (S) protein which initiates a host immune response. Both coronavirus SARS-CoV and SARS-CoV-2, the latter casing COVID-19 disease: invade the host cell from the binding between the S protein spike at the receptor protein angiotensin-converting enzyme 2 (ACE2), which is found on the surface membrane of host cells. This binding interaction results from an invasion process and requires S protein priming facilitated by the transmembrane serine proteinase 2 (TMPRSS2, known as epitheliasin) is present in the host cell. This proteinase was first identified in 1997 on human chromosome 21 by systematic exon-trapping experiments. It was found to be a multidomain type II transmembrane serine protease that cleaves the surface glycoprotein HA (haemagglutinin) of influenza viruses. This monobasic cleavage site is a condition for virus fusion and propagation. Also, it activates the fusion protein F of the human metapneumovirus and the spike protein S of the SARS-CoV. Therefore, TMPRSS2 is a potential target for drug design. Also, the SARS-CoV-2 viral genome encodes several non-structural proteins, such as 3-chymotrypsin-like cysteine protease (3CLpro). Therefore, this enzyme plays a role in coronavirus replication and is a proven drug discovery target for SARS-CoV and MERS-CoV.
Another important non-structural protein is RNA-dependent RNA polymerase (RdRp, also named nsp12) is an essential protein encoded in the genomes of all RNA-containing viruses with no DNA stage. This protein is central to coronaviral particles’ replication/transcription machinery and, therefore, a potential primary target for the antiviral drug remdesivir an adenosine analogue, which inserts into viral RNA chains, causing their premature termination). The RdRp catalyses the synthesis of the RNA strand, which complements a given RNA template, and the molecular mechanism for this process remains unclear. However, the RNA replication process involves a two-step mechanism:
- The initiation step of RNA synthesis commences at or near the 3′ end of the RNA template, facilitated by a primer-independent mechanism.
- This mechanism starts with adding nucleotide triphosphate (NTP) to the 3′-OH of the first initiating NTP. During the elongation phase, the nucleotidyl transfer reaction is repeated with subsequent NTPs to generate the complementary RNA product.
Combining zinc ions (Zn2+) and the zinc-ionophore pyrithione has been shown to inhibit nidovirus replication in cell culture. Further investigations are required for the use of zinc-ionophores as antiviral compounds. However, in vitro studies on the reversible inhibition of the RdRp by Zn2+ have provided a suitable research tool to gain more insight into the molecular details of (nido)viral RNA synthesis. It is important to reveal novel mechanistic differences between the RdRps of SARS-CoV and equine arteritis virus (EAV) in cell cultures.
The final non-structural protein papain-like protease (PLPro) from the human SARS-CoV consists of a cysteine protease located within the non-structural protein 3 (NS3) section of the viral polypeptide. The PLPro activity is required to process the viral polyprotein into functional, mature subunits. Also, PLPro cleaves a site at the amino terminus of the viral replicase part during viral protein maturation. In addition, PLPro possesses a deubiquitinating and deISGylating activity.
The SARS-CoV-2 enters the host cells; the viral genome is released as a single-stranded positive RNA (note positive-sense viral RNA genome and serves as a messenger RNA which can be translated into protein in the host cell). Consequently, the single-stranded positive RNA is translated into viral polyproteins using host cell protein translation machinery and is then cleaved into effector proteins by viral proteinases 3CLpro and PLpro. The PLpro also behaves as a deubiquitinase (a large group of proteases that cleave ubiquitin from proteins and other molecules) on specific host cell proteins. These include interferon factor 3 and NF-κB resulting in immune suppression. At this point, RNA-dependent RNA polymerase (RdRp) helps to make more viral genomic RNA containing negative-strand RNA to be used by RdRp.
The infection process initiated by coronavirus results from the interaction of viral S protein and ACE2 receptors on the host cell surface. The coronavirus binds to host cells through its trimeric spike glycoprotein, and therefore, this protein is a key target for potential therapies and diagnostic treatments. By cryo-electron microscopy, researchers have determined a 3.5-angstrom resolution structure of the SARS-CoV-2 trimeric spike protein. However, this S-protein can bind at least ten times more tightly than the corresponding spike protein of SARS-CoV to the common host cell receptor (ACE2). This binding has been shown for the RBD-specific monoclonal antibodies S230, m396 and 80R in SARS-CoV.
Therefore, this may be a reason why SARS-CoV-2 is of higher transmissible and contagious than SARS-CoV. Therapeutic agents are required to target the conserved proteins associated with SARS-CoV and SARS-CoV-2. Consequently, both RdRp and 3CLpro protease of SARS-CoV-2 share over 95% of sequence similarity with those of SARS-CoV. However, both these viruses demonstrate only 79% sequence similarity at the genomic level. On further investigation, it was found by applying sequence and homology modelling that both SARS-CoV and SARS-CoV-2 share a highly conserved receptor-binding domain (RBD) of the S protein with 76% in sequence similarity. Also, the PLpro sequences of SARS-CoV-2 and SARS-CoV are only 83% similar, and both viruses share a similar active site. To date, there are no SARS-CoV-2-specific antiviral agents.
Zoonotic Coronaviruses: Origins, Impacts, and the Race for Effective Vaccines
Coronaviruses belong to a large pool of viruses; some will cause illness in people, while others circulate among mammals and birds. However, it is unusual for animal coronaviruses to spread to humans and between humans. In the last decade, zoonotic coronaviruses have emerged in the human population, such as SARS-CoV-2 leading to COVID-19 disease, SARS and MERS. These betacoronaviruses lead to respiratory infection and, in some cases, gastrointestinal infections in humans. Moreover, the clinical range of these diseases varies from no symptoms or mild respiratory symptoms to severe. Further symptoms progress to pneumonia, acute respiratory distress syndrome, septic shock and multi-organ failure leading to death.
The acute respiratory infection SARS-CoV-2 was first identified in Wuhan City, Hubei Province, China, in December 2019. Since then, it has spread to over 180 countries around the World to become a pandemic which the WHO declared with a fatality rate of at least 2.3%. The respiratory tract infection caused by MERS-CoV was first identified in Saudi Arabia in 2012, and the case fatality rate was approximately 37%. These infectious diseases were compared to the acute viral respiratory tract infection caused by SARS-CoV, first known in the Guangdong province of Southern China in 2002. The SARS-COV epidemic affected 26 countries, resulting in more than 8000 cases and 774 deaths in 2003. However, there have been no reported cases since 2004, and the fatality rate was approximately 10%.
The World urgently needs a vaccine against SARS-CoV-2. Over the past decade, researchers have responded to other epidemics, including H1N1 influenza, Ebola virus disease (EVD, a viral haemorrhagic fever of humans and other primates caused by ebolaviruses), Zika (a member of the Flaviviridae family and originates from the Ziika Forest of Uganda, Aedes mosquitoes spread the virus) and SARS-CoV. These epidemics have produced a wealth of knowledge ranging from the discovery of pharmaceutical drugs to vaccine programmes. For example, a vaccine for H1N1 influenza was quickly developed because influenza-vaccine technology is well established. This monovalent H1N1 vaccine was available after the pandemic peaked and became a stand-alone vaccine incorporated into the seasonal influenza vaccines. However, vaccines for SARS, EVD and Zika followed a different path to H1N1 vaccine development. In this case, the SARS and Zika epidemics ended before vaccine development was completed.
Accelerating the development of vaccines to address epidemics led to the formation of the organisation known as the Coalition for Epidemic Preparedness Innovation (CEPI). This international non-governmental coalition comprises the Bill and Melinda Gates Foundation, Wellcome Trust, the European Commission and eight other countries Australia, Belgium, Canada, Ethiopia, Germany, Japan, Norway and the UK). CEPI aims to support the development of vaccines against five epidemic pathogens on the WHO list. CEPI will also support the development of a vaccine against the COVID-19 disease. Success in producing a vaccine would require support for development from viral sequencing to clinical trials and must be capable of large-scale manufacturing. In COVID-19, it is vital to manufacturing vaccines based on the DNA and RNA platforms, including recombinant-subunit vaccines.
The major advantage of vaccines made from RNA and DNA is that the processes do not require culture or fermentation and only use chemical synthetic processes such as those used for oncology vaccines. However, there are no approved RNA vaccines to date, but RNA vaccines have entered clinical trials. Developing a SARS-CoV-2 vaccine will impose different challenges; for example, could the virus S spike protein be a promising immunogen for protection? or would optimising the antigen design warrant an optimal immune response? Also, is it best to target the full-length protein or only the receptor-binding domain (RBD)?
However, past research on the SARS and MERS vaccines has raised concerns about worsening lung disease or facilitating antibody-dependent enhancement. These effects could be associated with type 2 helper T-cell (TH2) response, and therefore, it is best to carry out tests in suitable animal models. Furthermore, at present, the duration of immunity is unknown, and what would be the effect of single-dose vaccines on immunity?
In April 2020, a clinical trial involving about 510 volunteers will test a new vaccine called ChAdOx1 nCoV-19 for SARS-CoV-2 (COVID-19). This vaccine is an adenovirus vector (ChAdOx1) developed by researchers at the Oxford Jenner Institute. The vaccine contains the genetic sequence of this surface spike protein inside the ChAdOx1 construct. After vaccination, the surface spike protein of the coronavirus is produced, which primes the immune system to attack the coronavirus if it later infects the body.
Zoonotic diseases contribute to about 60-75% of emerging infectious diseases throughout the World, and more than 70% have supposedly originated in wildlife species. Bats are known to be a natural pool of viruses. These highly pathogenic viruses seriously threaten human health, especially coronaviruses responsible for SARS and MERS, the paramyxoviruses such as Nipah virus, hemorrhagic Ebola and Marburg filoviruses. The most recent pandemic linked to the coronavirus known as SARS-CoV-2, which originated in Wuhan, China, in December 2019, shared 96% identity with a bat-borne coronavirus at the whole-genome level. The emergence of MERS in 2012 and the ongoing COVID-19 pandemic have accelerated the interest in detecting coronaviruses of bat origin due to public health concerns. In addition to human-associated coronavirus, bats have also been implicated in the emergence and origin of swine acute diarrhoea syndrome (SADS), transmissible gastroenteritis virus (TGEV) in pigs and porcine epidemic diarrhoea (PED). Therefore, it is possible that bat-borne coronavirus could pose a considerable threat to human health and food production. Researchers have discovered seven new coronaviruses detected in bats in Myanmar. However, none of the viruses was closely related to SARS-CoV, MERS-CoV, or SARS-CoV-2.
Article Update – Decoding COVID-19: Structural Insights and Global Response Efforts
The article “COVID-19: A Race Against Time—Understanding Pandemic Speed” explores the structural and genetic complexities of the virus responsible for COVID-19, known as SARS-CoV-2. It elucidates the virus’s structure, detailing its spherical form with a diameter of 80 to 160 nanometres and surface ‘crown’ projections that facilitate its entry into host cells. This encapsulation is crucial for the virus’s replication process, leveraging its positive-sense RNA to produce essential proteins and initiate infection within the host.
The taxonomy of coronaviruses categorizes them into alpha coronaviruses and beta coronaviruses, with the latter group including significant human pathogens like MERS-CoV, SARS-CoV, and SARS-CoV-2. The article provides a foundational understanding of how the virus’s spike proteins bind to host cell receptors, a pivotal step in the viral infection process.
Moreover, the text outlines the epidemiological trajectory of COVID-19, tracing its origins back to Wuhan, China, in December 2019. Initial containment efforts, the identification of the novel virus, and subsequent global spread are chronicled, highlighting the swift international response aimed at curbing the pandemic’s impact. Insights into the virus’s receptor-binding domains suggest similarities between SARS-CoV-2 and other coronaviruses, enhancing our understanding of its transmission capabilities.
Finally, the article discusses ongoing research and development efforts aimed at confronting the pandemic, including vaccine development and therapeutic interventions. These efforts underscore the critical need for a nuanced understanding of SARS-CoV-2 to effectively inform public health strategies and mitigate the pandemic’s effects. This comprehensive examination not only offers a detailed look at the scientific aspects of the virus but also emphasizes the global challenge posed by COVID-19 and the concerted efforts required to overcome it.
Disclaimer
The information presented in this article is intended for educational and informational purposes only. It reflects the understanding and knowledge available as of the date of publication (19 April 2020) and may not reflect the most current developments in COVID-19 research or clinical practice.
Open Medscience does not provide medical, diagnostic, or treatment advice. The content should not be used as a substitute for professional medical judgement, advice, diagnosis, or treatment. Always consult a qualified healthcare provider with any questions regarding a medical condition or before undertaking any healthcare-related action.
This article references scientific and clinical data that were available during the early stages of the COVID-19 pandemic. Given the rapidly evolving nature of SARS-CoV-2 research, readers are advised to consult updated resources and guidelines issued by reputable health authorities such as the World Health Organization (WHO), the UK National Health Service (NHS), and the Centres for Disease Control and Prevention (CDC).
The inclusion of data, references, or studies within this article does not constitute endorsement by Open Medscience of any particular method, treatment, or diagnostic tool. Any mention of experimental or investigational therapies is solely for the purpose of scientific discussion.
Open Medscience makes no representations or warranties of any kind, express or implied, regarding the accuracy, completeness, or reliability of the information provided. The authors and publisher accept no liability for any loss or damage arising from reliance on the content herein.